Introduction
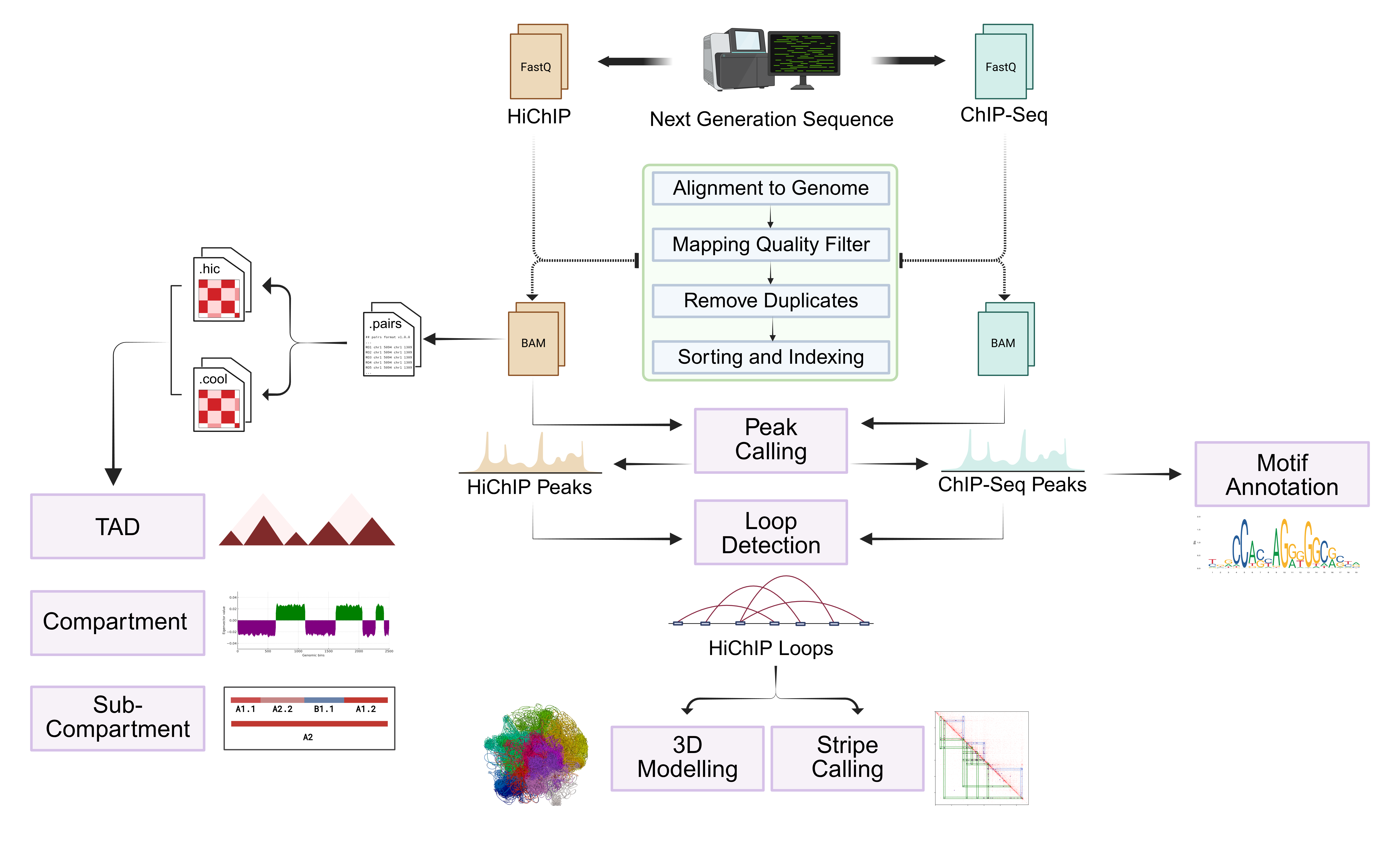
dcHiChIP is a modular and reproducible Nextflow-based pipeline designed for high-throughput analysis of chromatin architecture using HiChIP data. The pipeline supports a wide range of tools for mapping, loop calling, peak annotation, chromatin feature extraction, and 3D modeling.
The pipeline includes integrated modules for:
- Mapping & filtering of HiChIP/ChIP-seq reads using
BWA,SAMtools, andMACS3 - Loop calling using
MAPSand conversion to BEDPE format - Feature extraction with
cooltools,HOMER,gStripe, andCADLER - Multi-resolution compartment and TAD analysis
- 3D modeling of genome architecture using
MultiMM
Working Test Cases
The pipeline supports three fully documented test cases with representative input formats and figure overviews.
👉 Visit Working Test Cases for more.
Module Parameters
Each pipeline module supports customizable parameters such as resolution, CPUs, input formats, and modelling levels.
👉 Visit Module Parameters for full details.
Contact Us
For help, questions, or feedback, feel free to reach out.
👉 Visit Contact Page
Citation
If you use dcHiChIP in your research or publication, please cite the following manuscript (in preparation):
ContributorsAgarwal, A., Al-Bkhetan, Z., Plewczyński, D. dcHiChIP: A comprehensive Nextflow-based pipeline for Multi-Scale Analysis of Chromatin Architecture from HiChIP Data.