dcHiChIP supports three primary use cases, designed to accommodate the most common experimental scenarios involving HiChIP and ChIP-seq data.
Each case is automatically detected and managed by the pipeline based on input parameters and file formats provided in the configuration file.
Case 1: HiChIP FASTQ Only
This mode processes raw HiChIP paired-end FASTQ files and performs end-to-end analysis, including alignment, peak calling, loop detection, and 3D modeling.
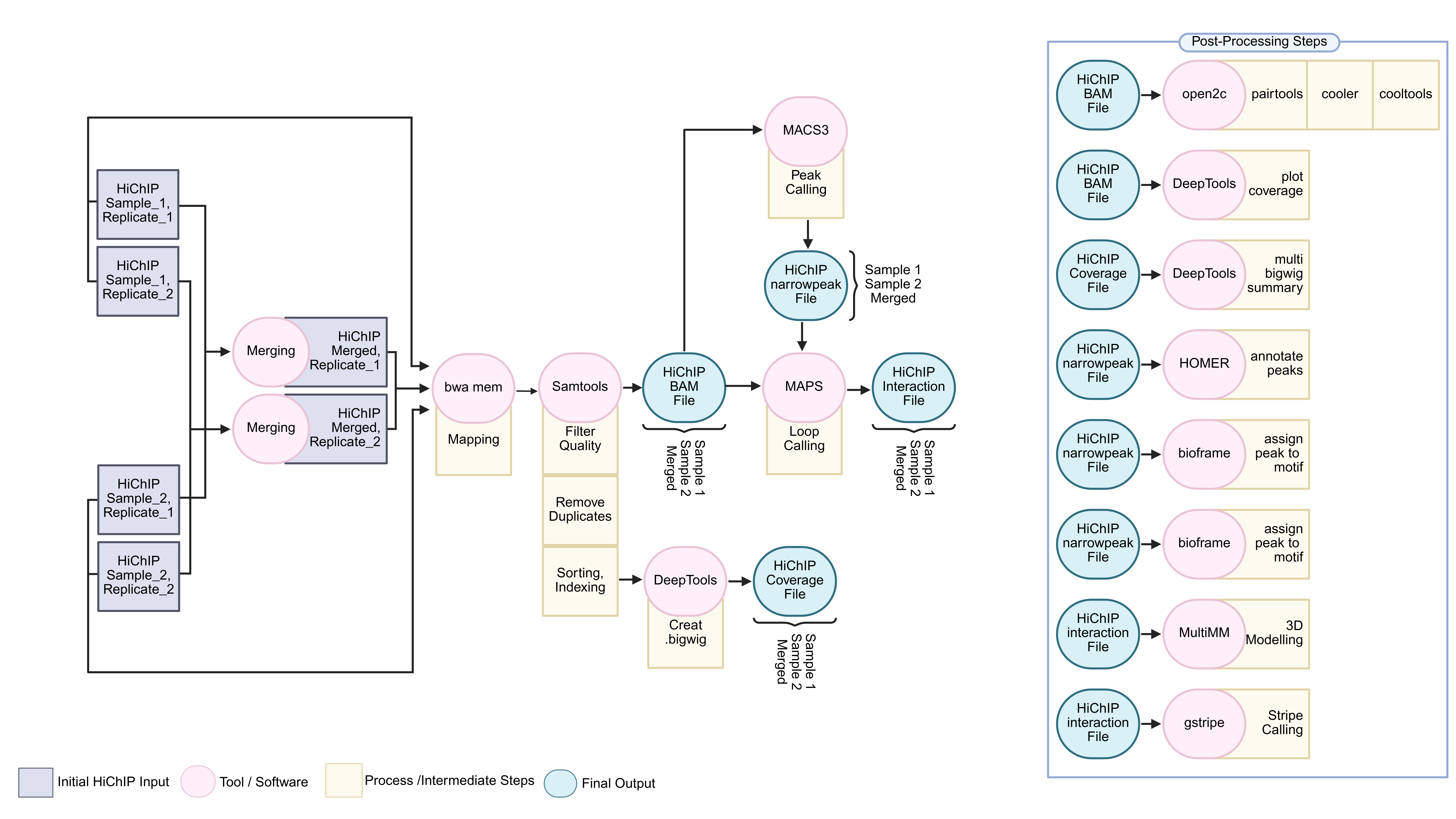
Use this Samplesheet when you do not have ChIP-seq data (neither raw nor peak files).
| id | group | hichip_r1 | hichip_r2 | chipseq_r1 | chipseq_r2 | narrowpeak |
|---|---|---|---|---|---|---|
| S-1 | S1 | ./dchichip/SAMPLE1_R1.fastq.gz | ./dchichip/SAMPLE1_R2.fastq.gz | |||
| S-2 | S1 | ./dchichip/SAMPLE2_R1.fastq.gz | ./dchichip/SAMPLE2_R2.fastq.gz |
Notes:
- The last column (
chipseq) & (narrowpeak) must be left empty. - Pseudo-ChIP-seq peaks will be automatically generated from the HiChIP data.
Case 2: HiChIP FASTQ + Pre-Processed ChIP-Seq Peaks
In this mode, the user provides raw HiChIP FASTQ files along with pre-processed ChIP-seq peaks in narrowPeak format. This bypasses the need for peak calling from HiChIP reads.
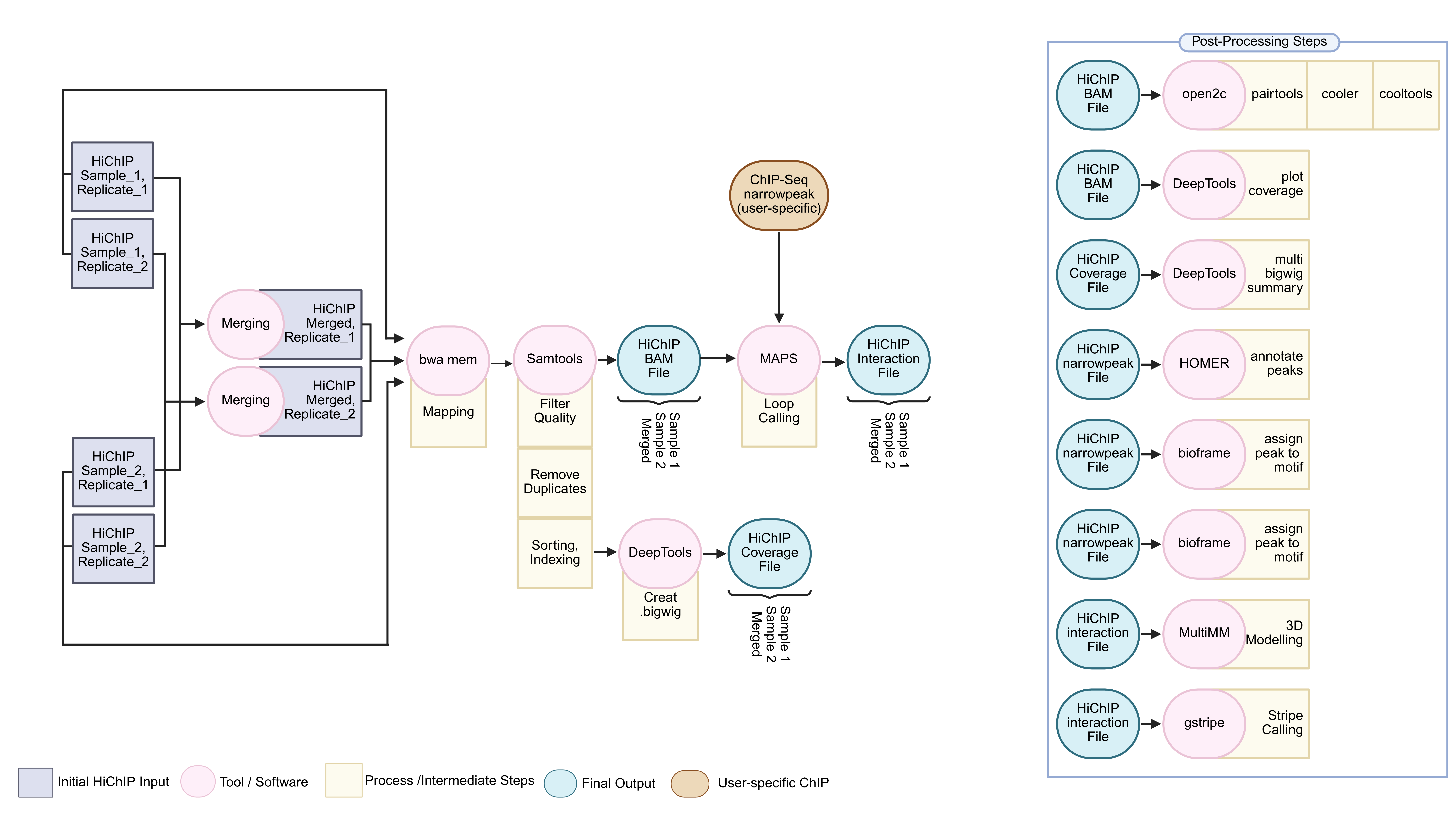
Use this format when you have pre-processed ChIP-seq peak files in narrowPeak format.
| id | group | hichip_r1 | hichip_r2 | chipseq_r1 | chipseq_r2 | narrowpeak |
|---|---|---|---|---|---|---|
| S-1 | S1 | ./dchichip/SAMPLE1_R1.fastq.gz | ./dchichip/SAMPLE1_R2.fastq.gz | ./chip/SAMPLE1.narrowpeak | ||
| S-2 | S1 | ./dchichip/SAMPLE2_R1.fastq.gz | ./dchichip/SAMPLE2_R2.fastq.gz | ./chip/SAMPLE2.narrowpeak |
Case 3: HiChIP FASTQ + ChIP-Seq FASTQ
This configuration performs joint processing of both HiChIP and ChIP-seq FASTQ files. ChIP-seq reads are aligned, and peaks are called independently to guide HiChIP loop detection.
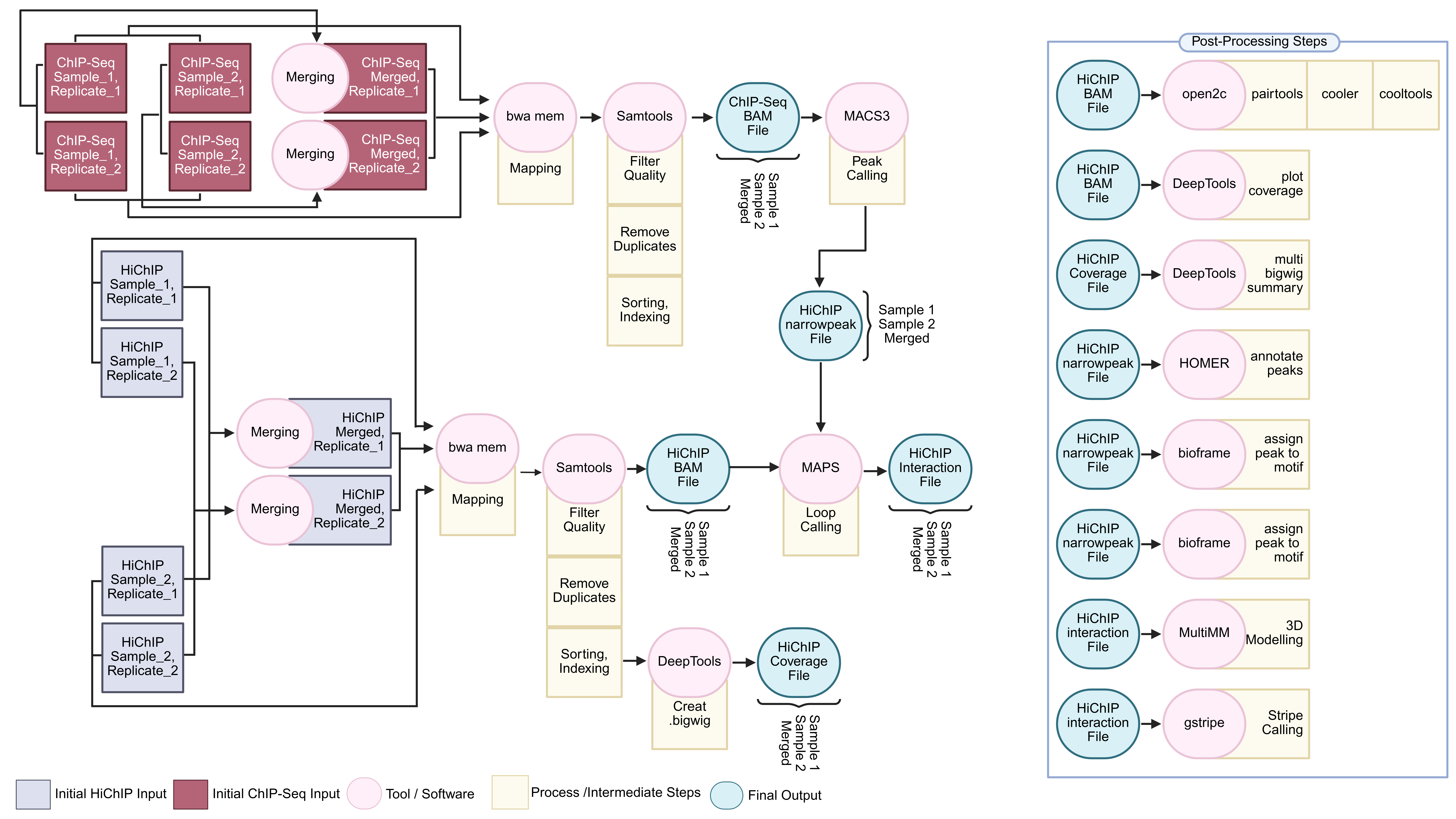
Use this format when you have raw ChIP-seq data and wish to process it within the pipeline.
| id | group | hichip_r1 | hichip_r2 | chipseq_r1 | chipseq_r2 | narrowpeak |
|---|---|---|---|---|---|---|
| S-1 | S1 | ./dchichip/SAMPLE1_R1.fastq.gz | ./dchichip/SAMPLE1_R2.fastq.gz | ./chipseq/SAMPLE1_R1.fastq.gz | ./chipseq/SAMPLE1_R2.fastq.gz | |
| S-2 | S1 | ./dchichip/SAMPLE2_R1.fastq.gz | ./dchichip/SAMPLE2_R2.fastq.gz | ./chipseq/SAMPLE2_R1.fastq.gz | ./chipseq/SAMPLE2_R1.fastq.gz |
Notes:
- ChIP-seq input files must correspond to the same sample and replicate scheme.
- Peak calling will be automatically performed using MACS3.